Step 1
Create your own Google Spreadsheet file
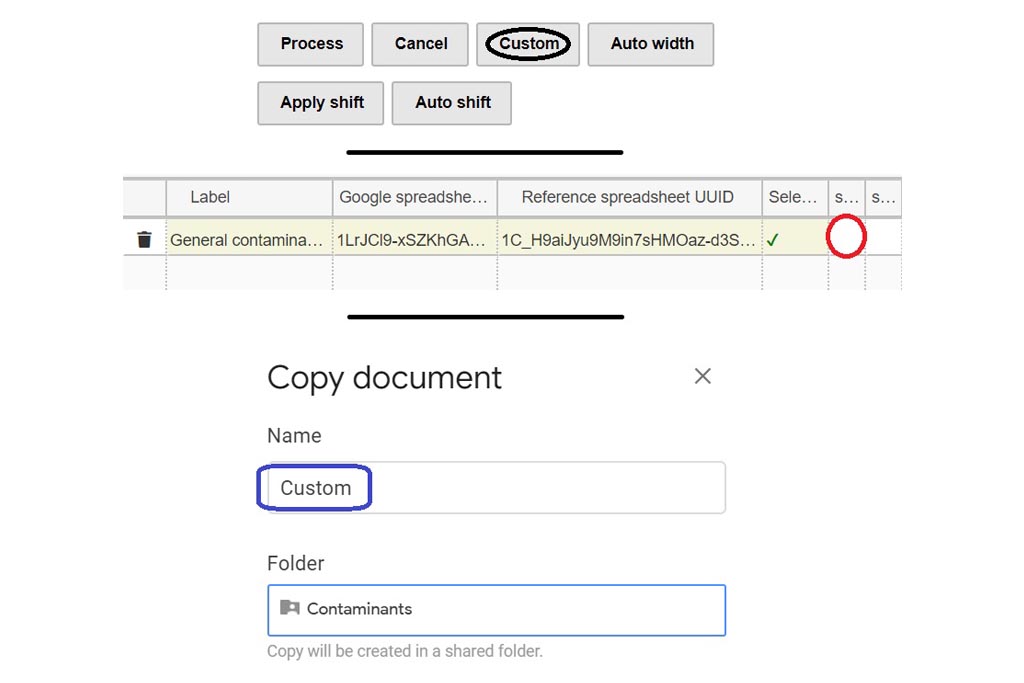
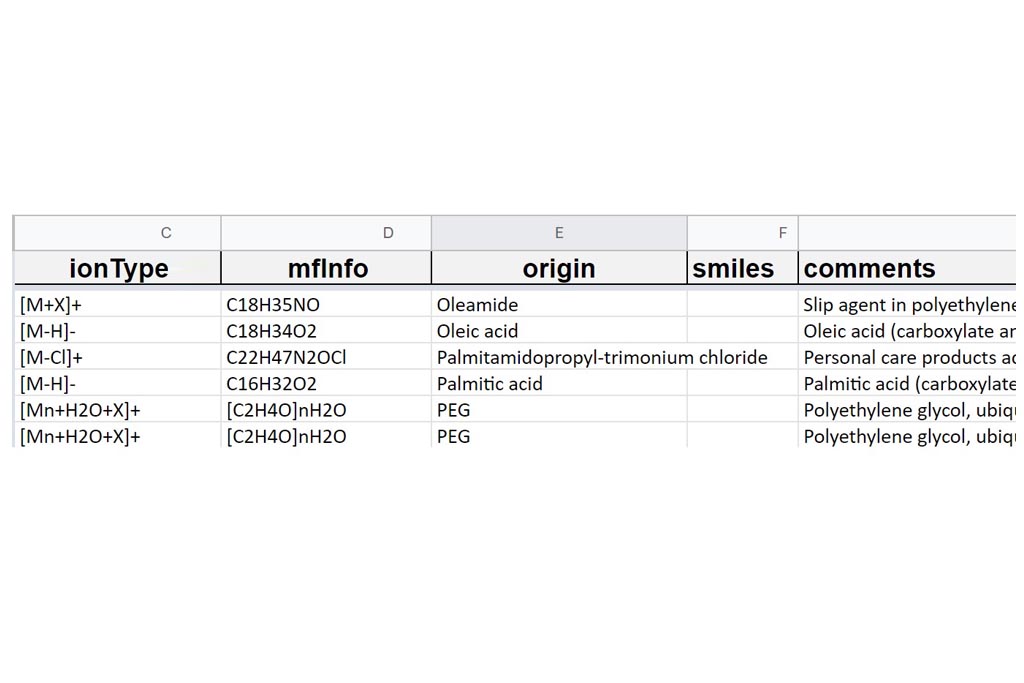
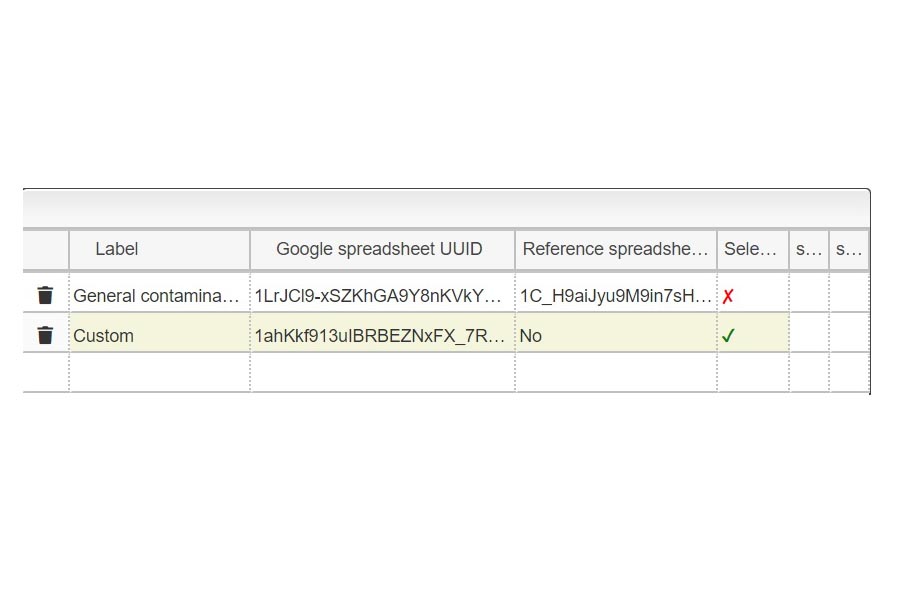
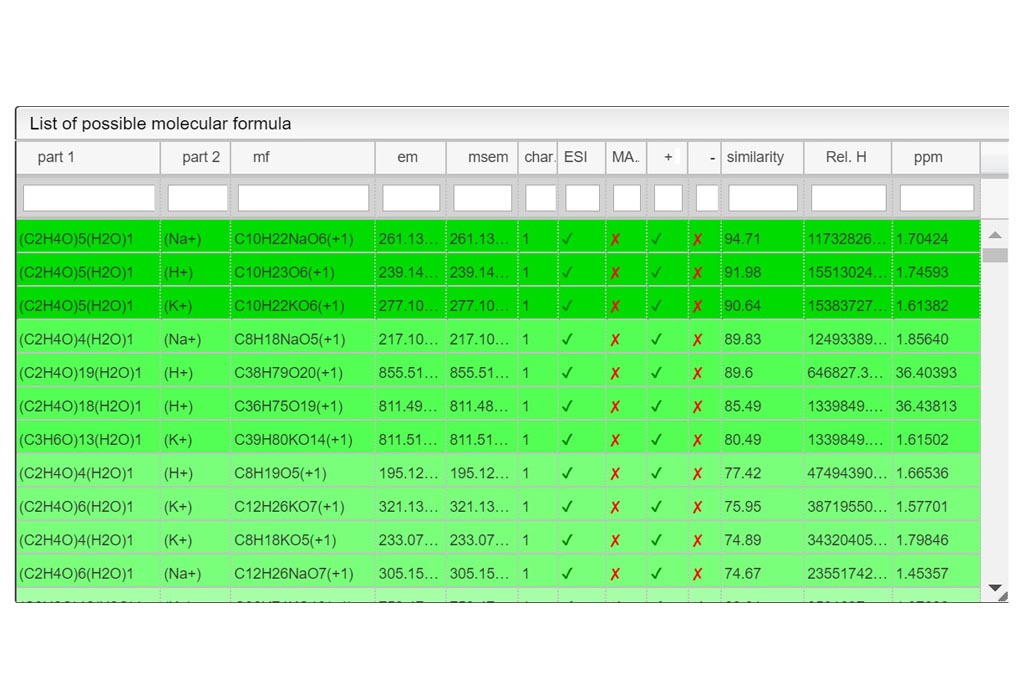
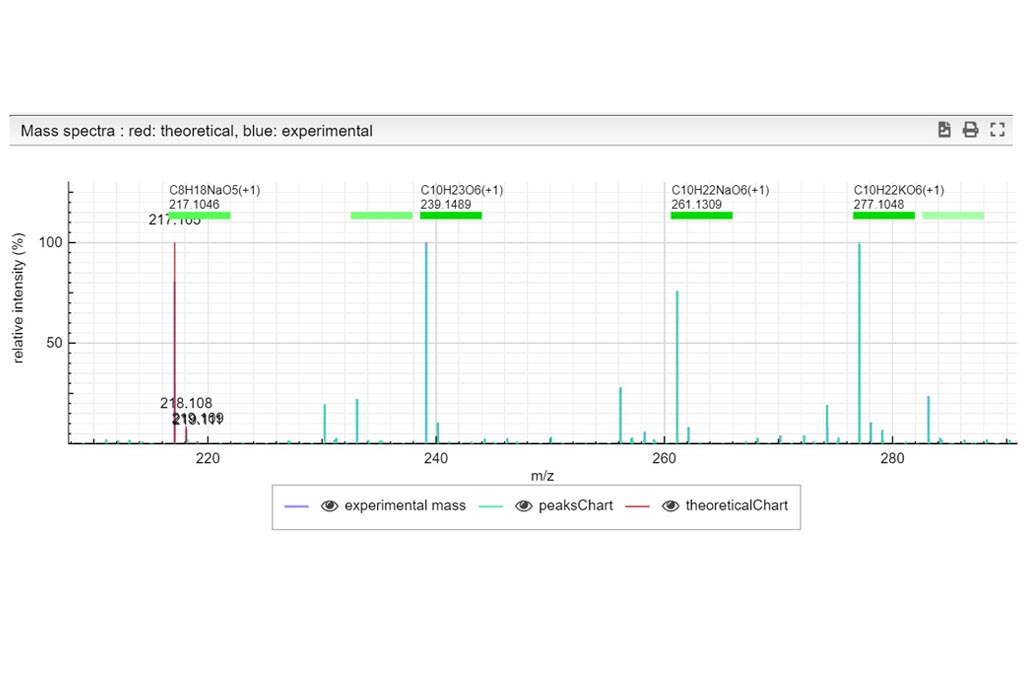
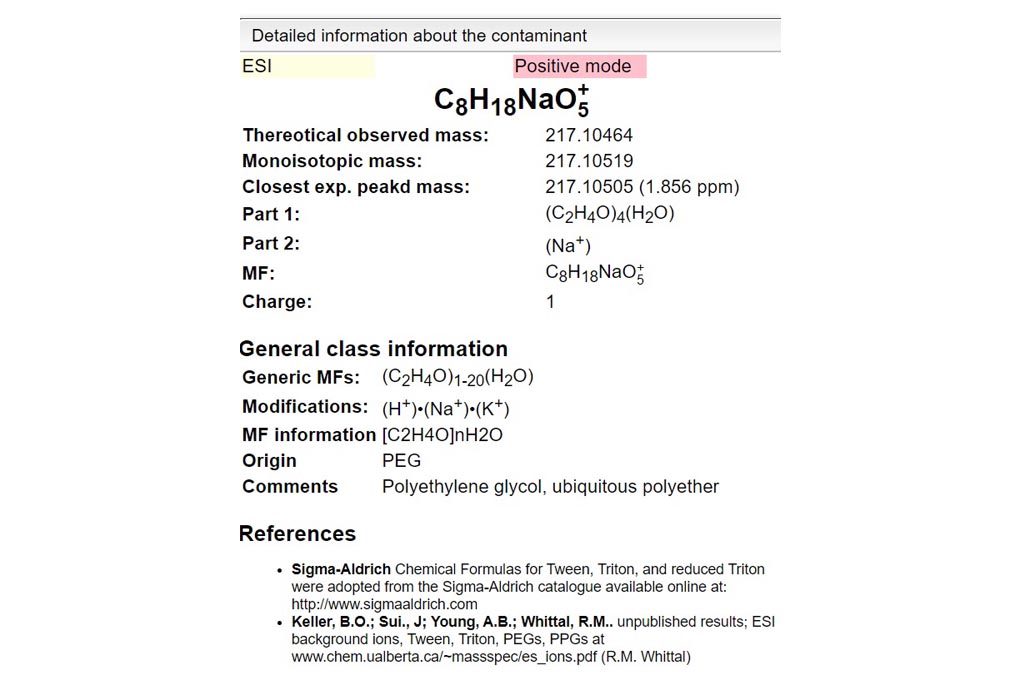
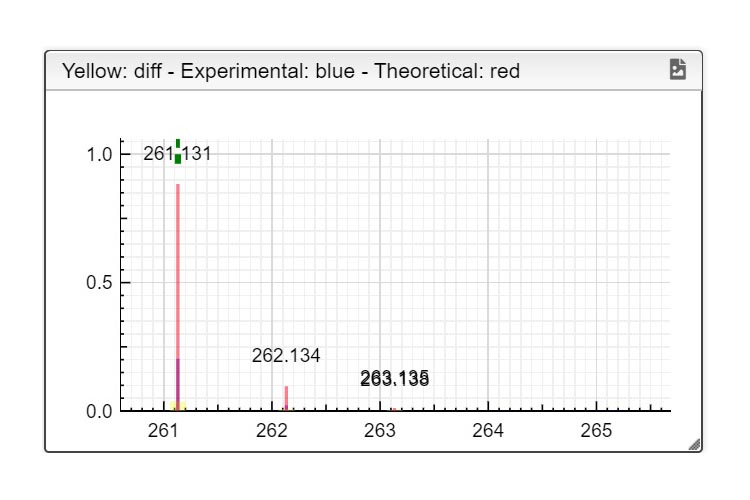
First you must click on Custom button signaled in the figure as a black oval (1). In the custom module you have to open the existing google spreadsheet signaled in the figure as a red circle (2). Once open, go to file and make a copy, change the name signaled as a blue rounded rectangle (3) and folder. Then, you can delete raws and start your own DB from scratch by entering your own compounds.