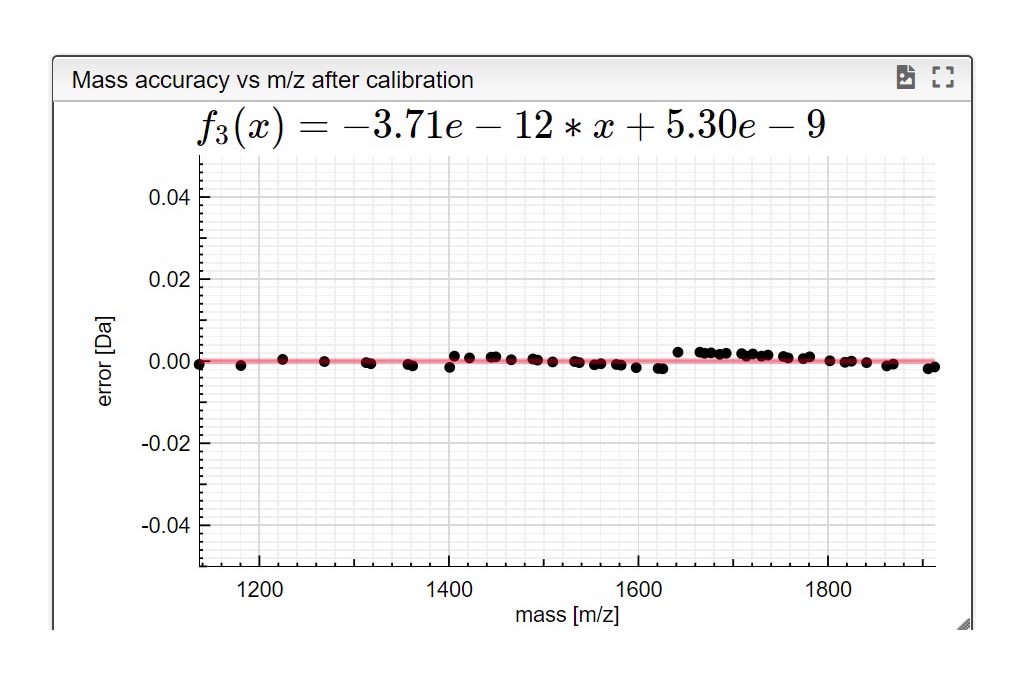
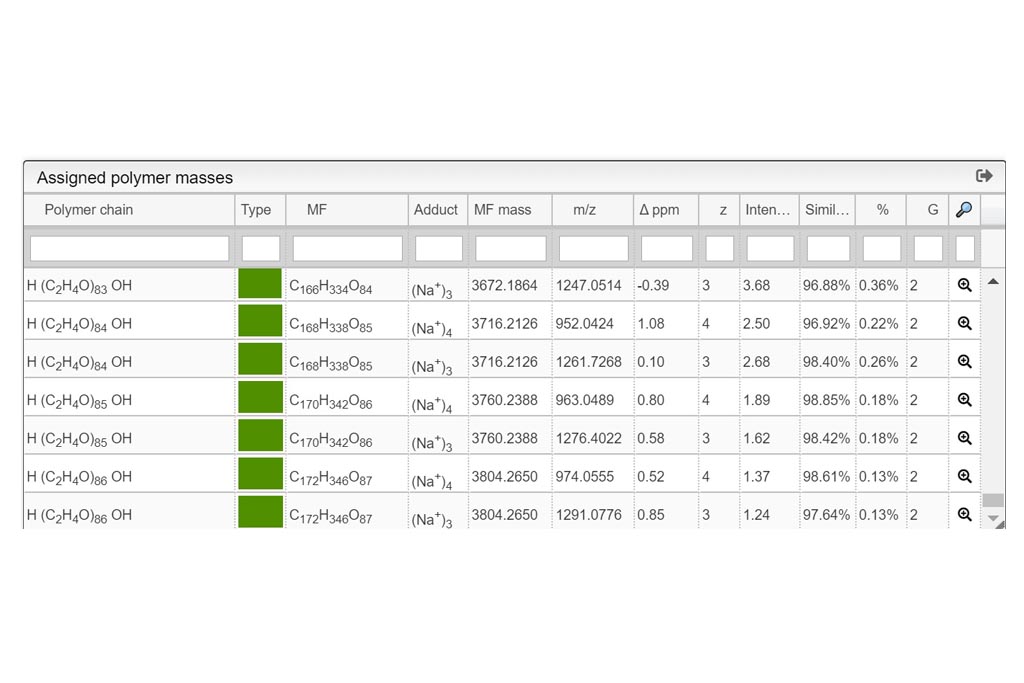
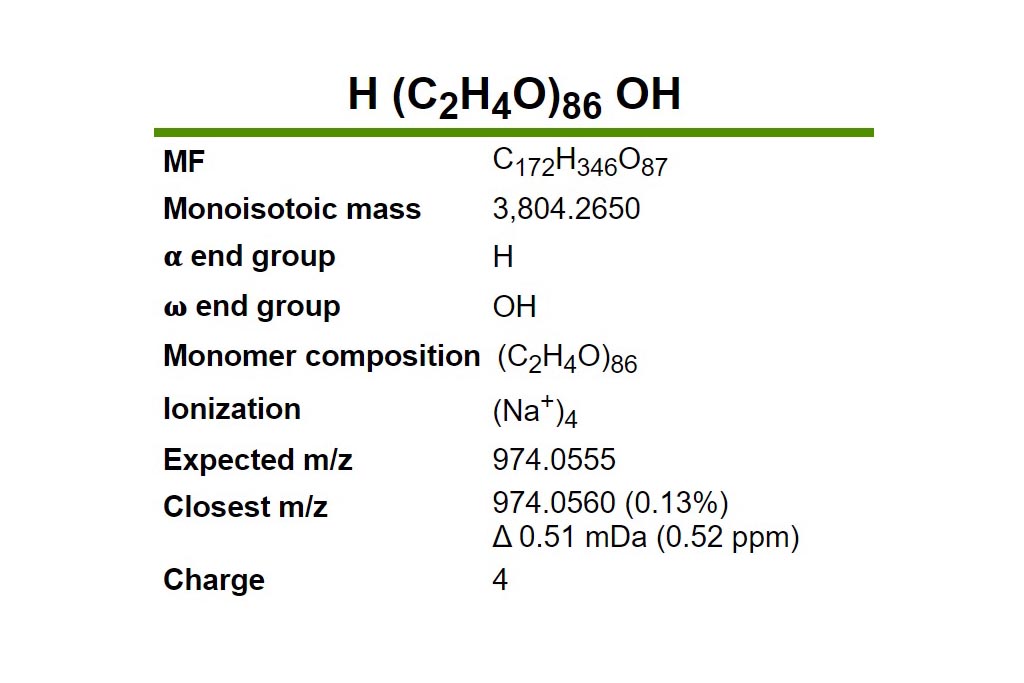
Resuts 5
Kendrick View
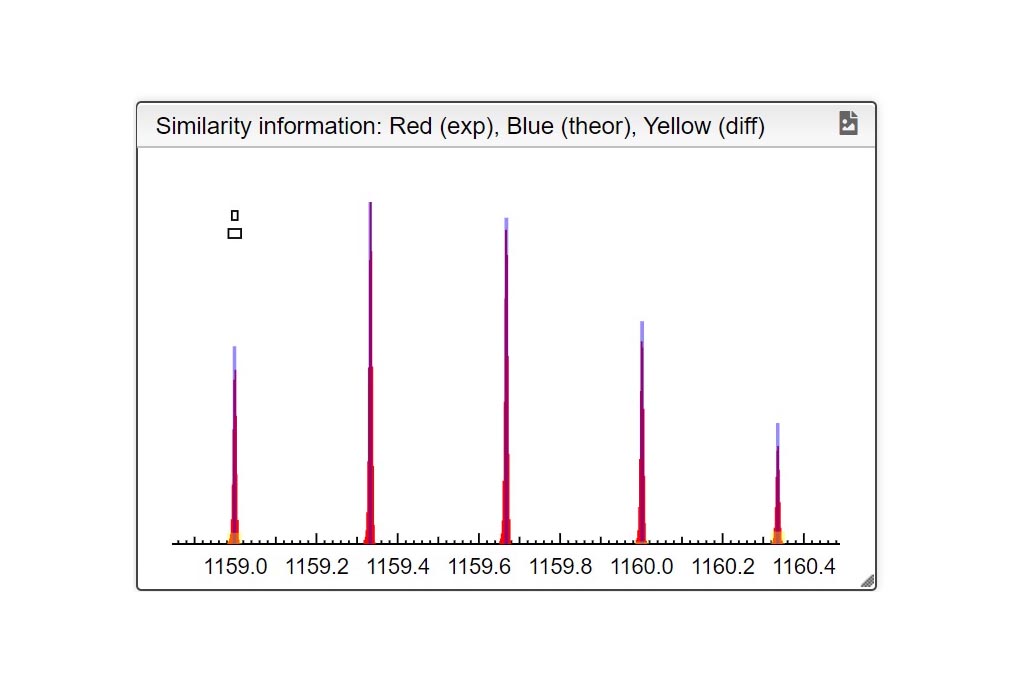
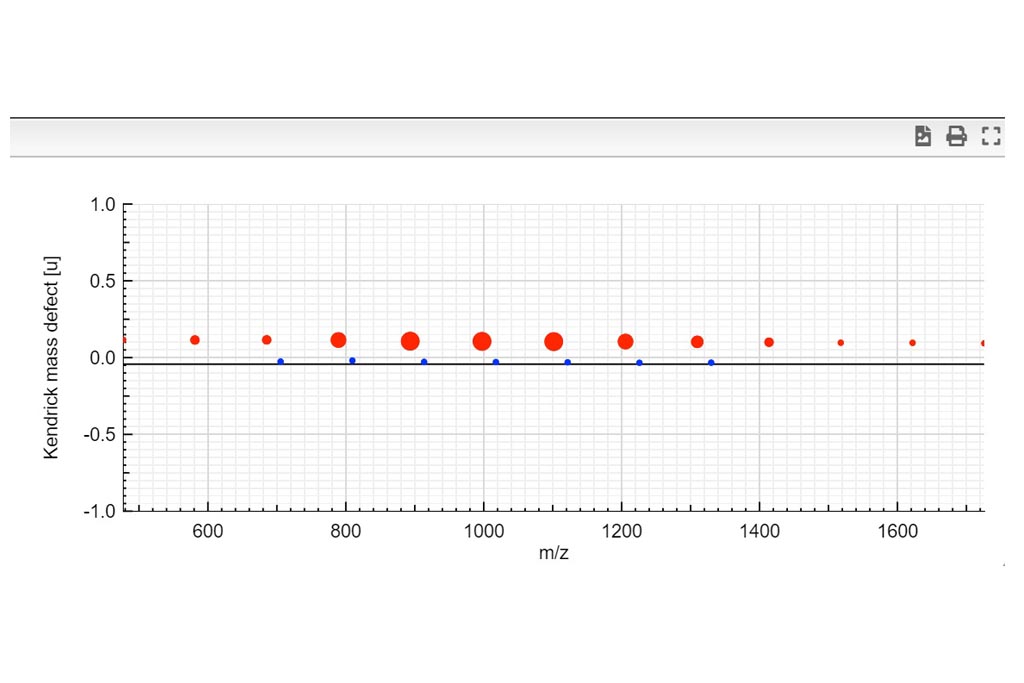
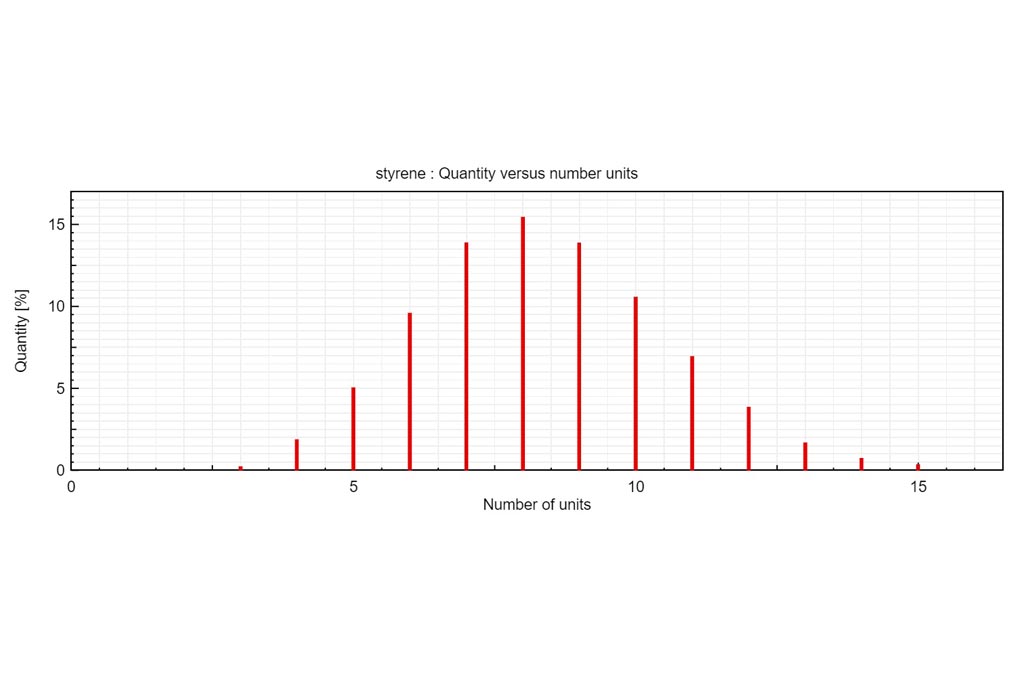
Kendrick mass defect plots were integrated in MSPolyCalc from the perspective of unravelling spectra complexity by offering a correlative approach. Two options are available. On one hand, the m/z KMD plot is determined from experimental m/z values (referring to [M+Na]+ for example). It yields a representative picture of the experimental mass spectrum, with different split distributions of same chemical composition but different ionization adduct. On another hand, a simplified KMD plot, named “deconvoluted” KMD plot can be plotted. In this case, mass defects are calculated from the MF masses, thus referring to the neutral species [M], and neglecting the contribution of the ionization adducts. Kendrick Tab is interactive and moving the mouse cursor over the plots controls what the Info Box displays.