STEP 3
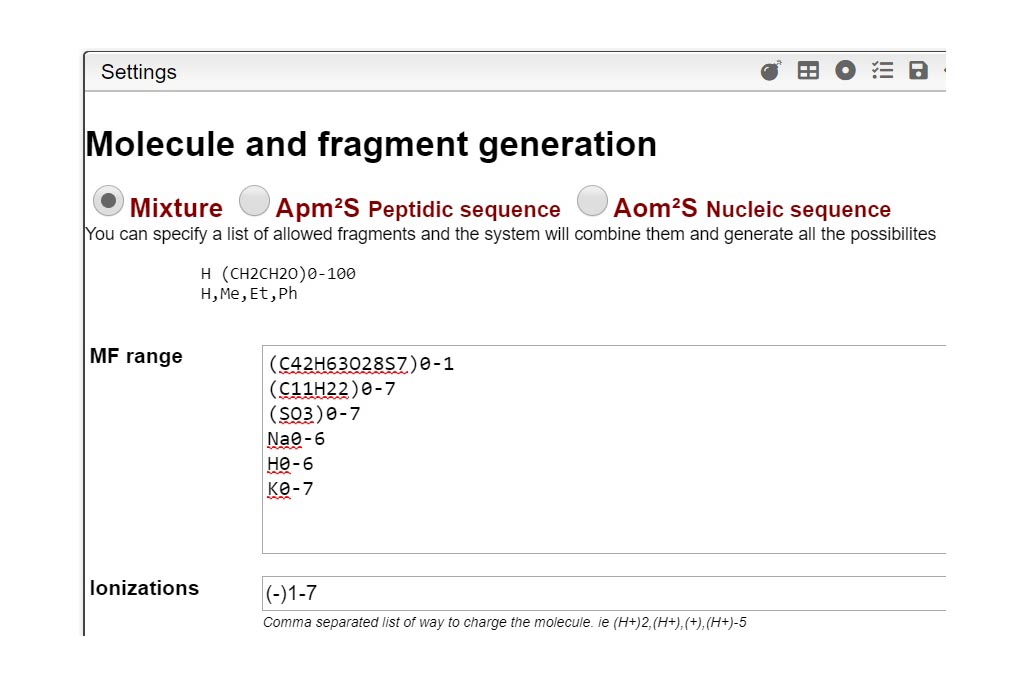
Molecular Formula range selection and description
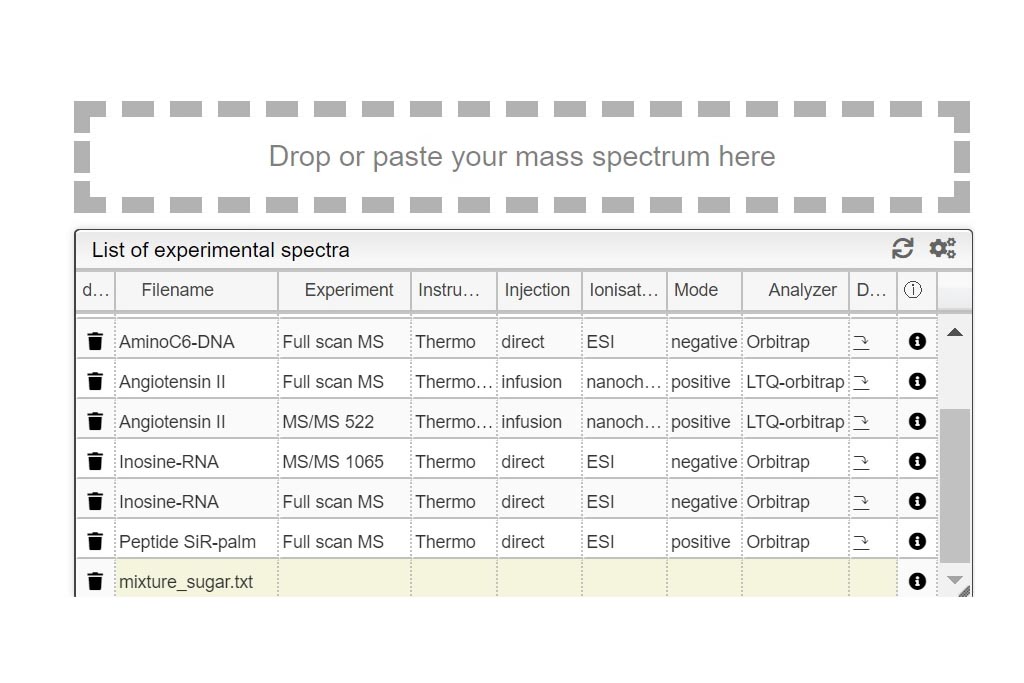
This step is critical for a good mixture calculation. You need to define all possible “pieces” that you want to find in your experiment. Like in a puzzle, the solver is expected to put pieces together in a logical way. Let’s think about the sugar examples shown here, where one lipophilic central core should be present but you want to probe also for absence, written (C42H63O28S7)0-1). This core tends to bind between 0-7 chains (C11H22SO3) in our case. During the experiment, some labile SO3 groups may have been lost during ionization so the chain could be split in two (C11H22)0-7 and (SO3)0-7. Each of the sulfate group is negatively charged so you should specify it by indicating it this way [-1- -7]. Additionally, the molecule could also by cationized with H,Na or K positively charged. Mixture will combine all these possibilities and generate their corresponding molecular formulas. Finally mixture will subsequently try to matched theoretical to the experimental spectra. The list of all abbreviations that can be used to indicate the common functional groups is found at the top right corner of the Settings window.