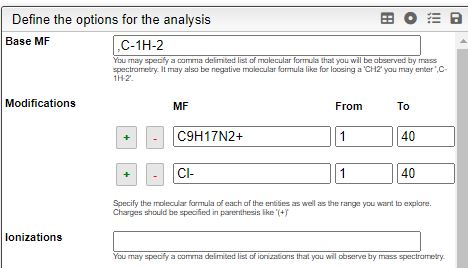
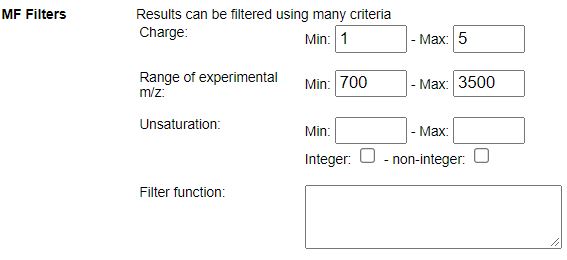
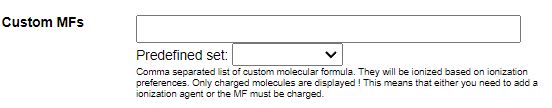
STEP 1
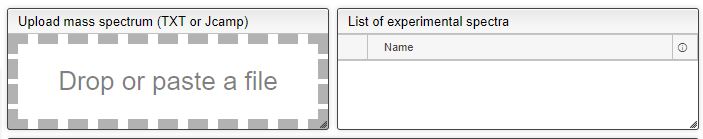
Importing Experimental data
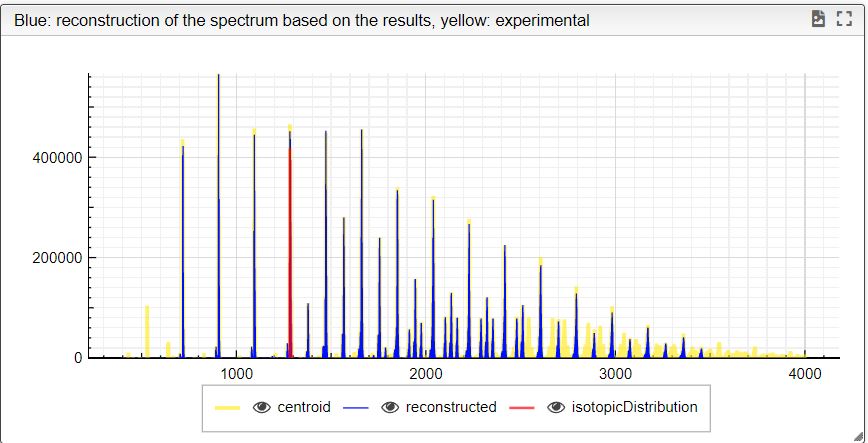
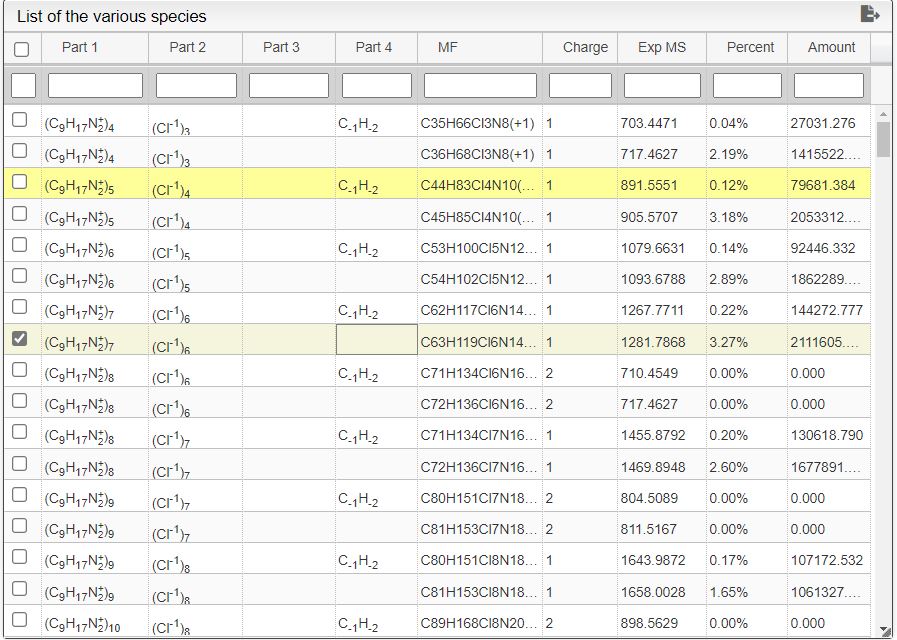
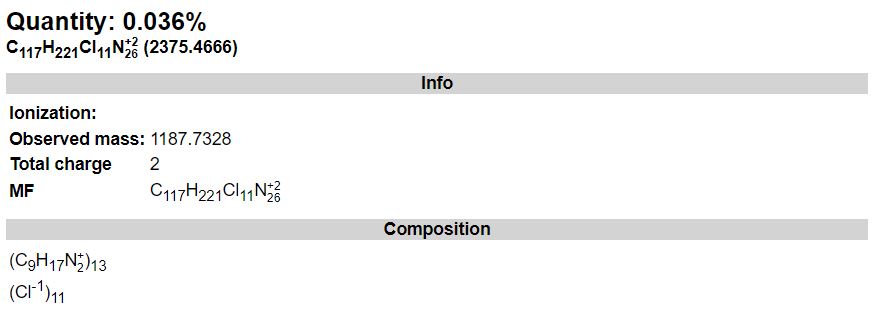
This tool can be either used in a LIMS or stand-alone. In the stand-alone mode you should either drag/drop your experimental spectrum as a tab-delimited text file or copy paste it (CTRL-V) while moving you mouse over the drop zone. The list of available spectra will be displayed in the table and you may click on one of them to display it.