Preferences
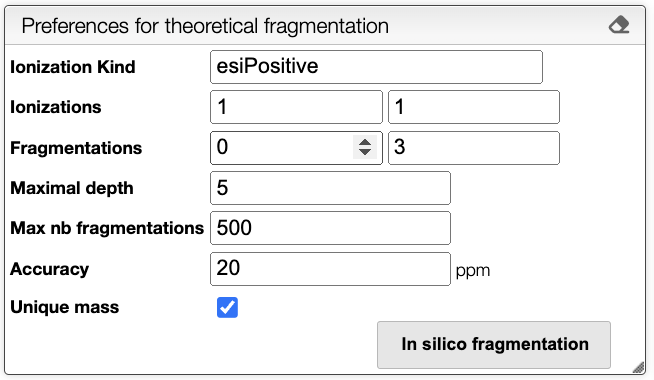
To compute the in silico fragmentation, the user needs to provide parameters in the left panel.
It is possible to choose the following parameters:
- Ionization Kind: Kind of ionization, currently only
esiPositiveis supported. - Ionizations: Range of ionizations to consider. The default is
1-1. - Fragmentations: Number of fragmentation mask to apply. The default is
0-3. - Maximum depth: Maximum depth of the fragmentation tree. The default is
5. - Max nb fragmentations: Maximum number of reactions to consider. The default is
500. - Accuracy: Accuracy in the mass in ppm. The default is
20. - Unique mass: If checked, only unique masses are considered. The default is
true.
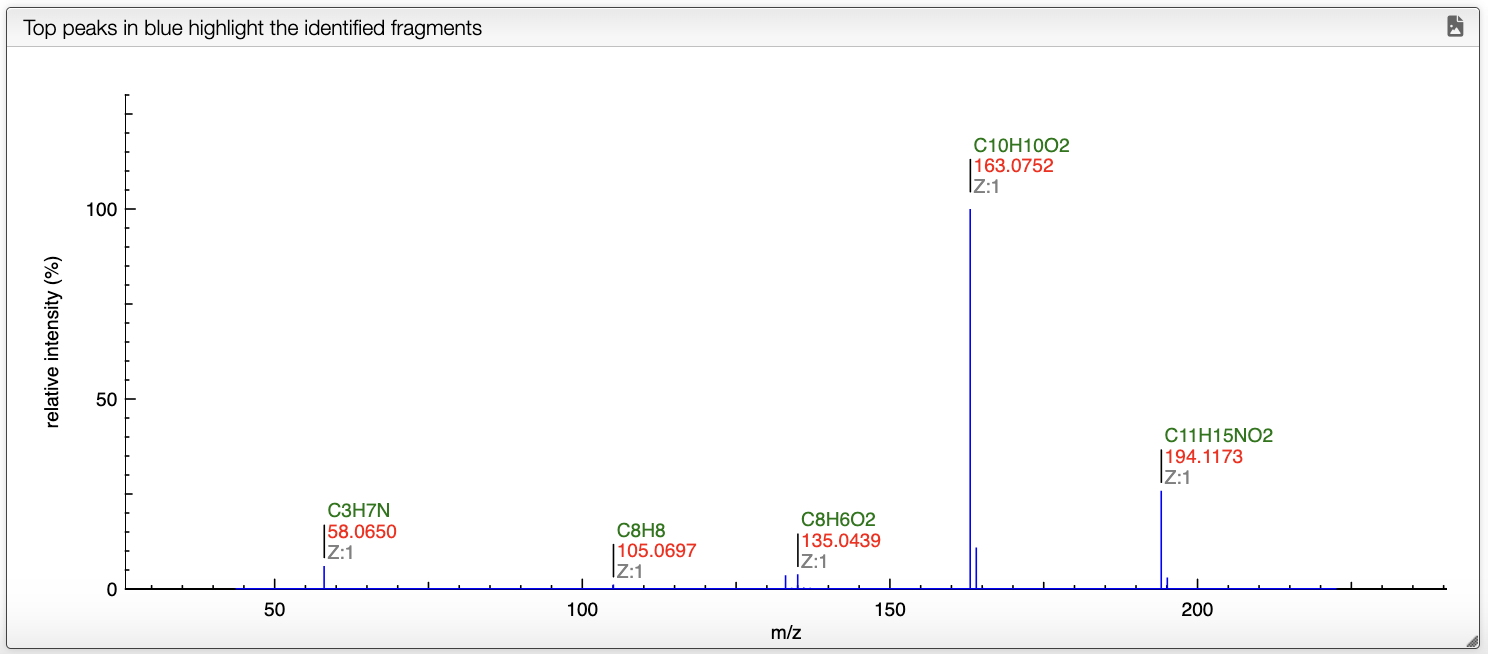
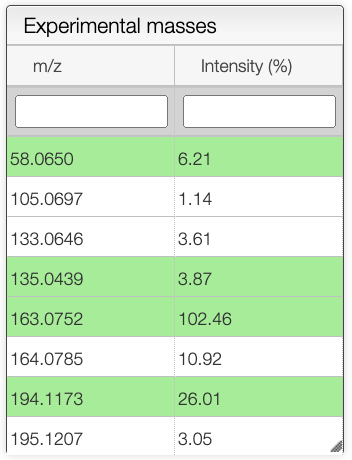
Once all the parameters are set, the user can select a spectrum in the spectrum panel. If there is no spectrum, the user can upload one by drag and dropping a file in panel that is below the spectrum panel.
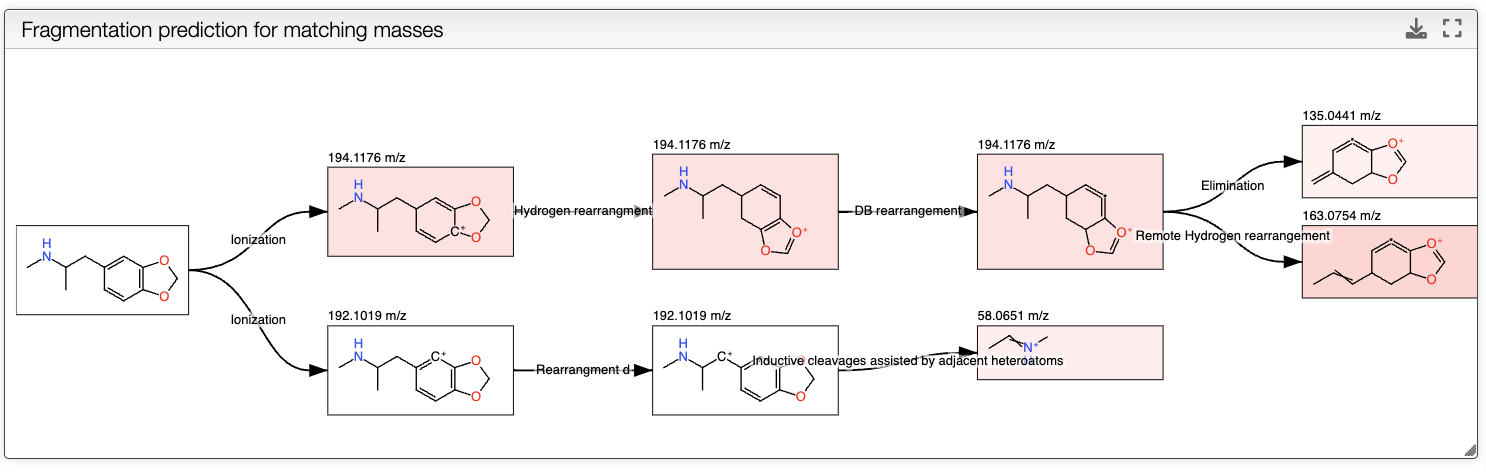
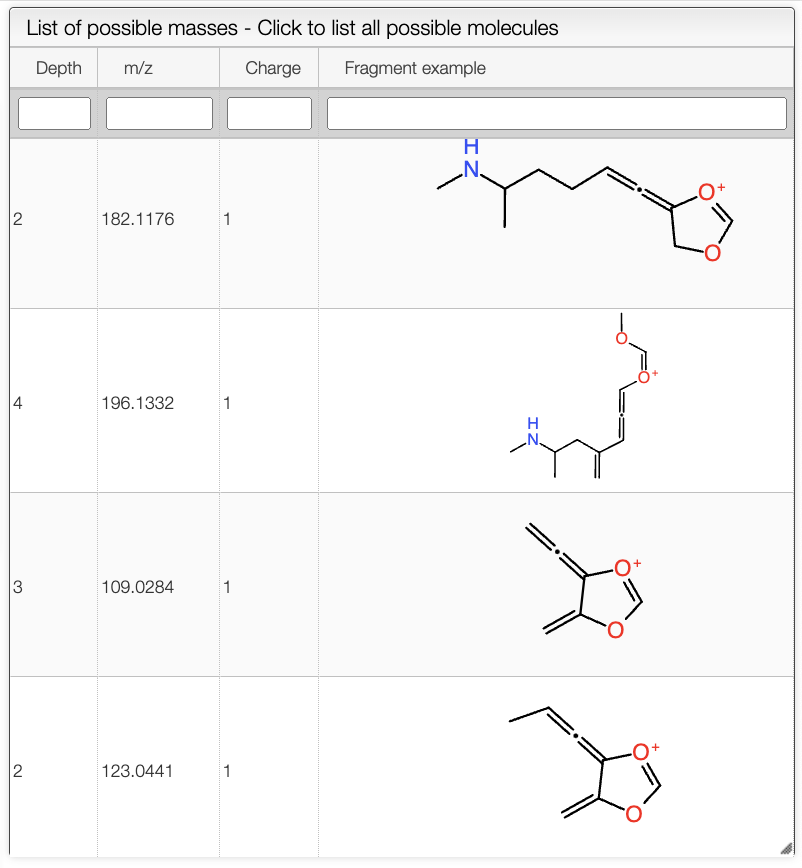
When a spectrum is selected, it will automatically be show in the bottom panel, and processed to label the peaks with the molecular formula associated. This annotation is based on the molecular formula of the molecule drawn in the bottom left panel.