Pubchem (Pubchem tab)
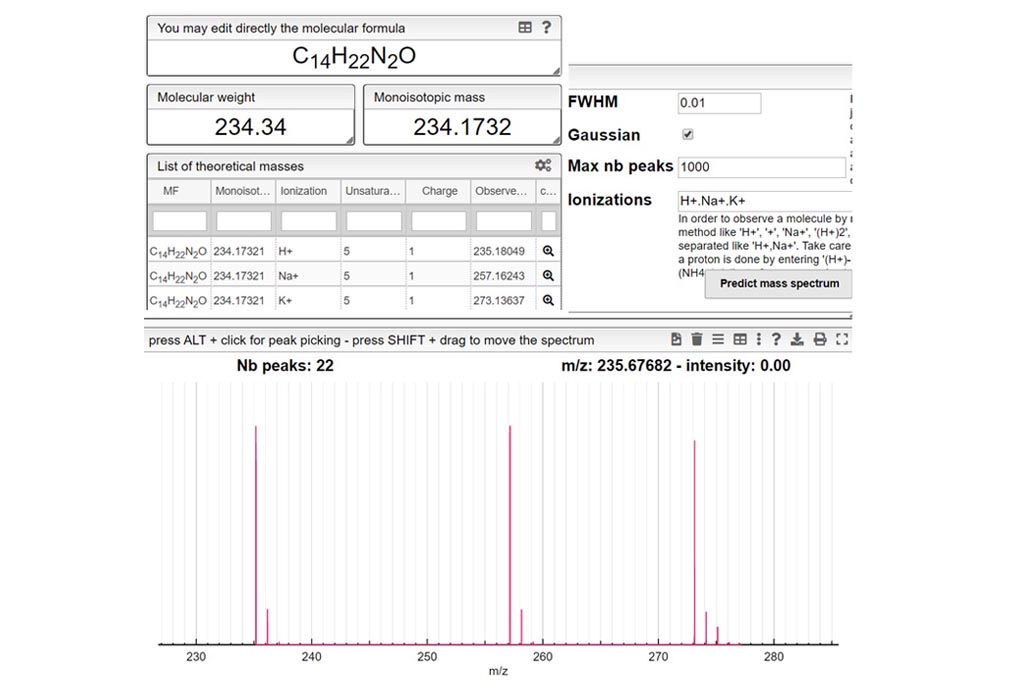
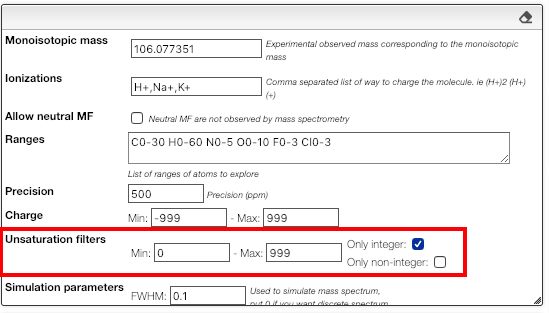
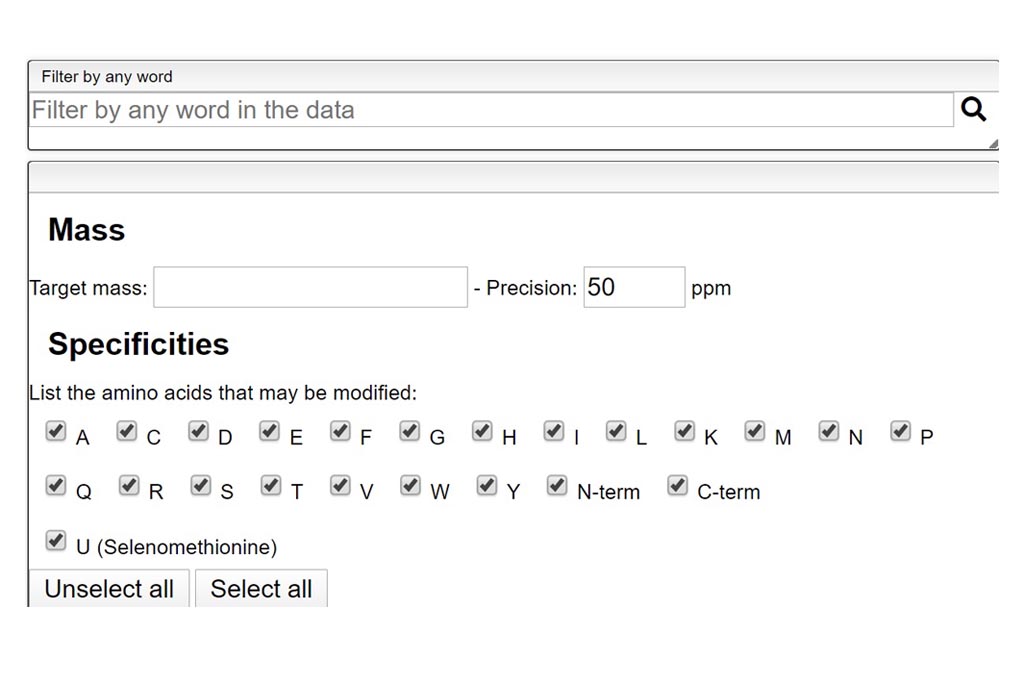
This tool allows to find a molecular formula, even if we don’t know the range of atoms that compose the molecule. A few assumptions are made: the charge of the non ionized molecule is null, there are at least 5 molecules that have the same MF and the molecule does not break apart.
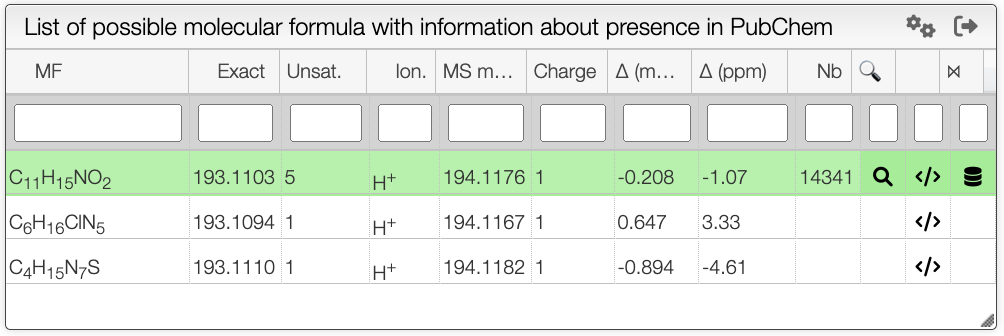
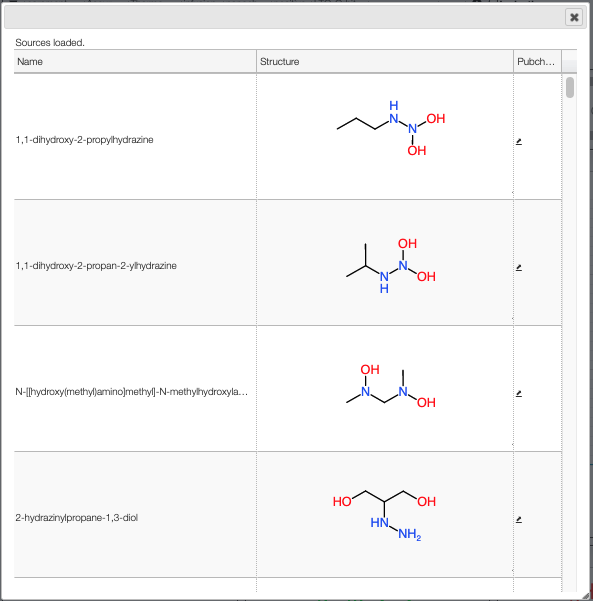
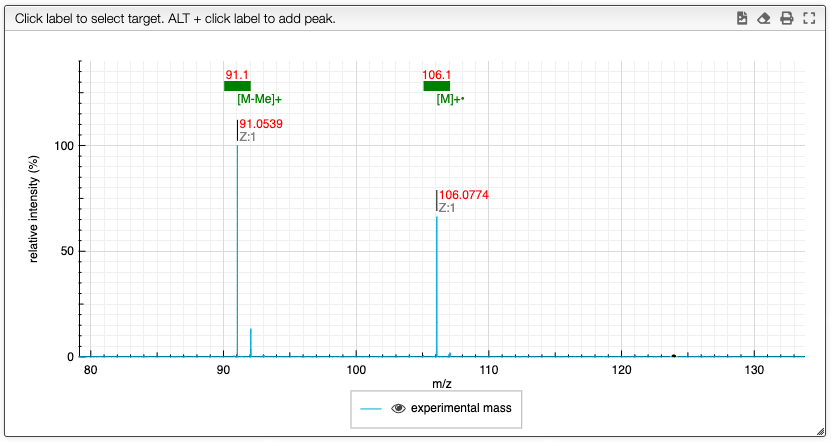
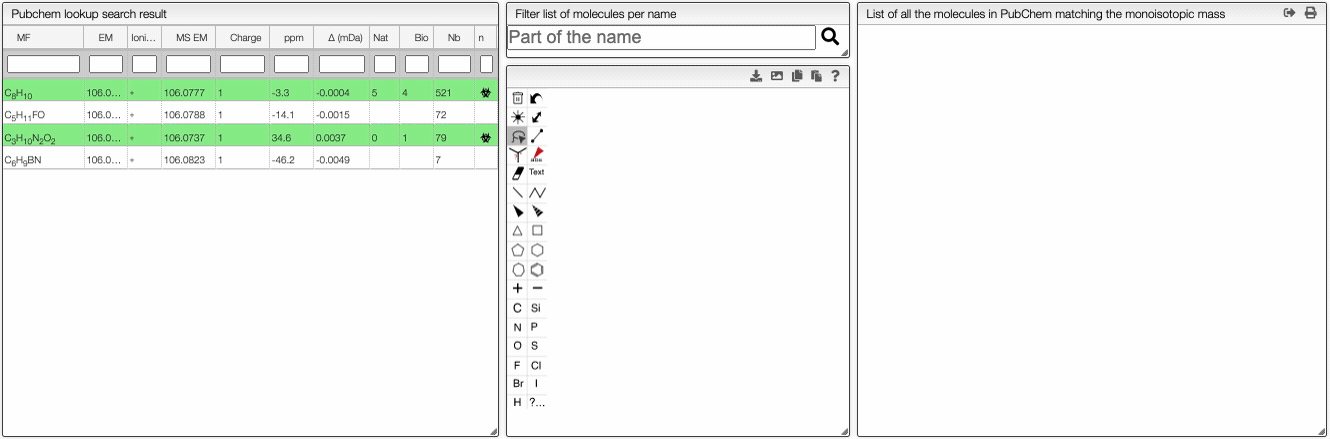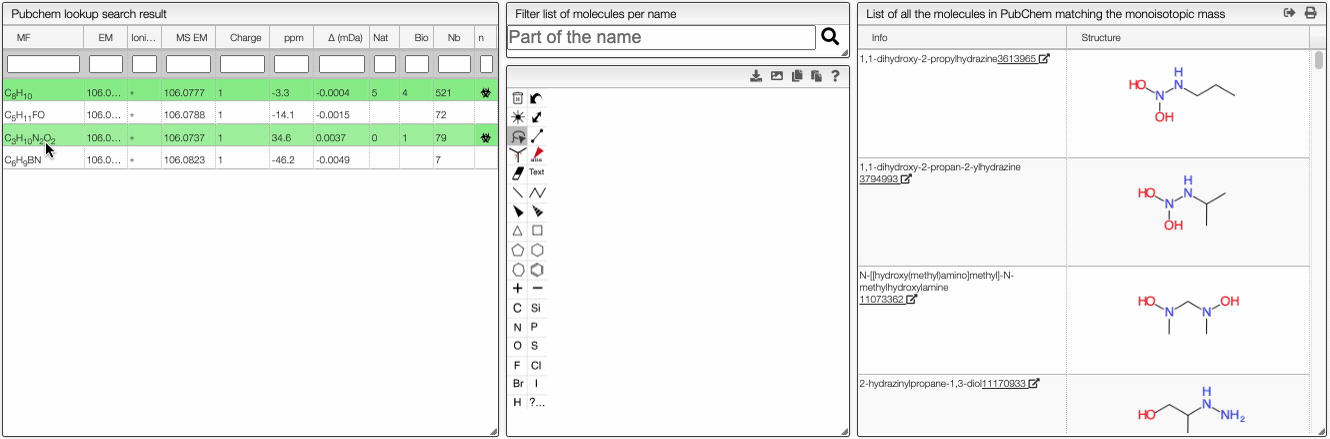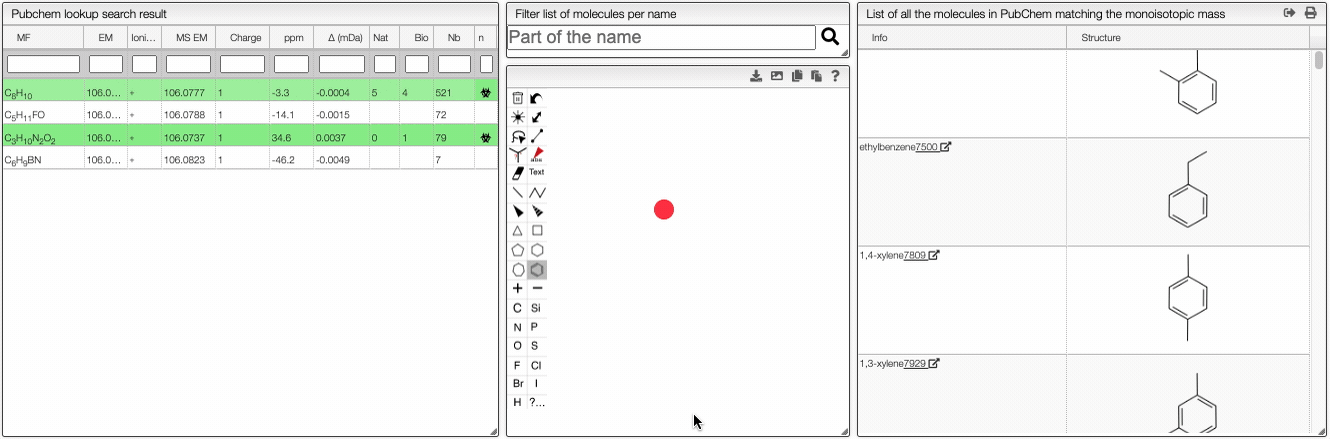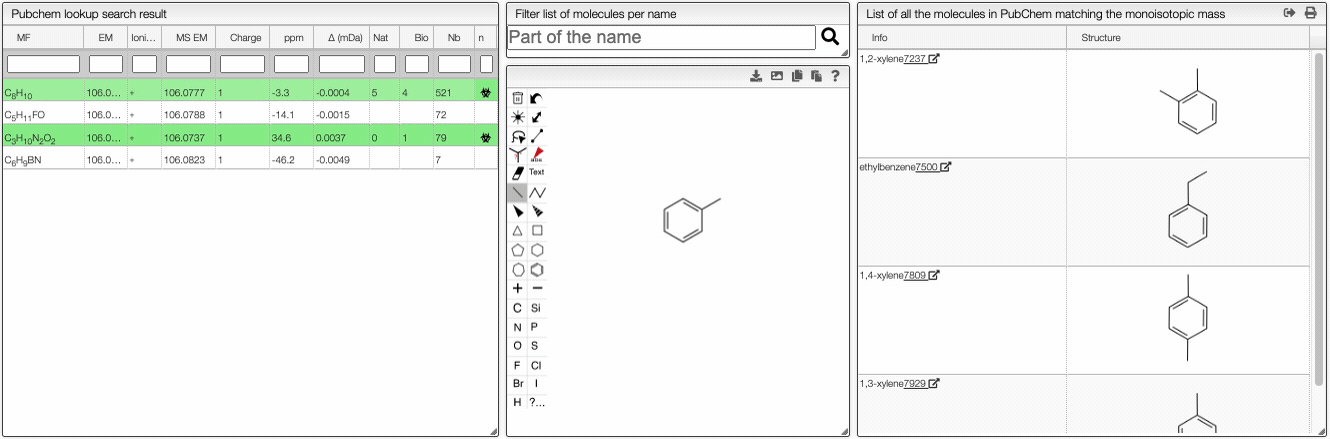
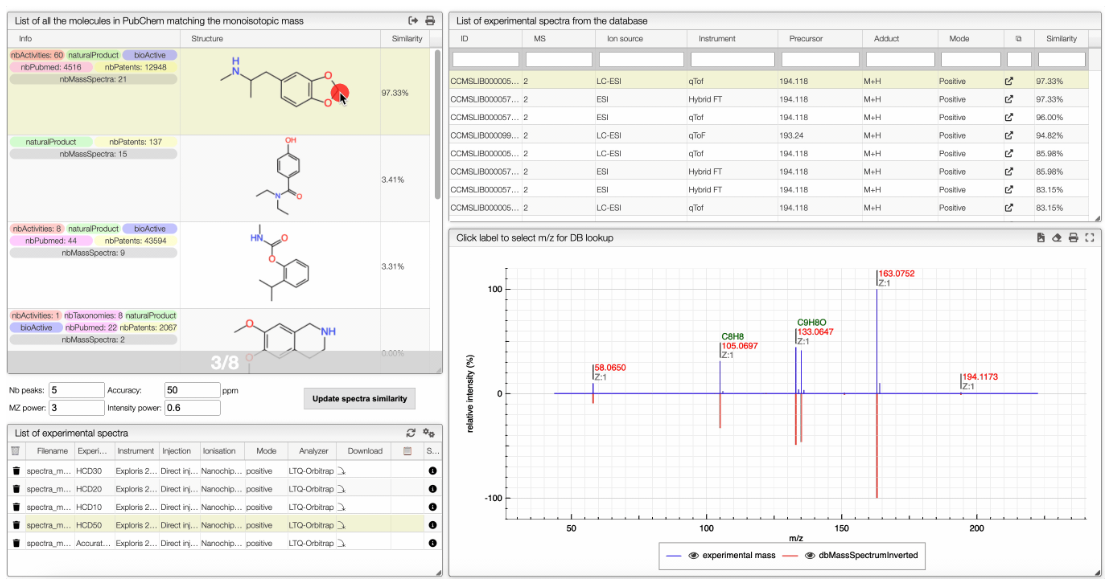
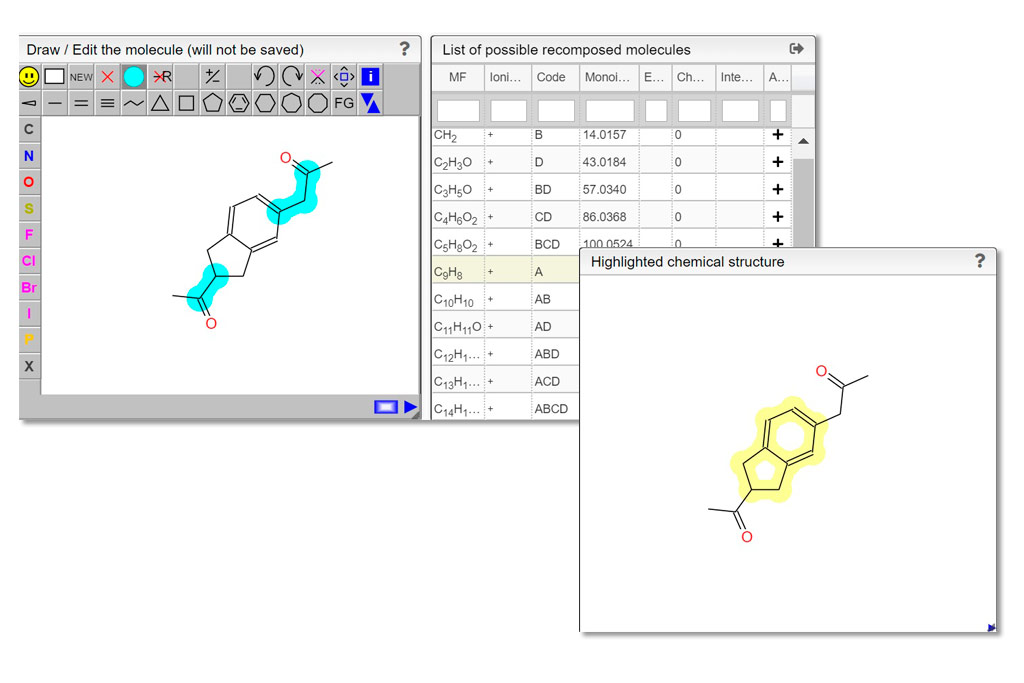
When the spectrum is loaded, you can click on a peak and the system will show a list of the possible molecular formula for a given ionization and accuracy. On the table you can see in green the molecular formula that are registered in the PubChem database. If you click on a formula, you can see at the right the list of isomers. In the draw panel, you can make a substructure search or a name search within the list.
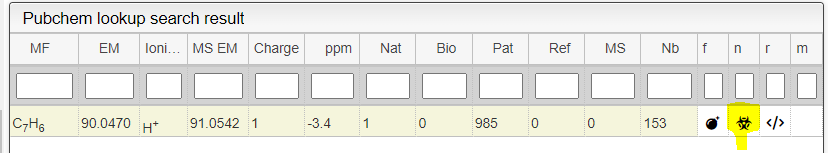
In the table you can also see the number of natural product (Nat) and bioactive (Bio) molecules with the same molecular formula. If you click on the icon, you can see a table with the publication in PubMed related to the natural or bioactive product. Moreover, using MeSH terms, you can filtrate the results in the search bar at the top.
The results of the search are displayed in a table. The table includes the structure of the compounds as well as tags such as the number of taxonomies, activities, patents, mass spectra… It also shows if the compounds is a natural product or not.
On the Detail information panel, you can find more information about the compound. On the top of the panel, there are the tags associated with the number of mass spectra, number of patents… Below, there is the structure of the compound without stereochemistry, the number on the right indicates the number of stereoisomers found in the database. By clicking on it, you will find all the structures as well as where it comes from.
Pubmed (Medline): In this section, you will find all the publications associated with the compound. By clicking on the title of the publication, you will be redirected to the Pubmed page. The number on the right of each title indicates the number of times that this compounds has been described in the publication. A green color indicates that the publication can be interesting. This allows the user to quickly find the most relevant publications. The three dots next to the number allow the user to see the abstract of the publication. The abstract includes the mesh terms.
Activities: Activities show additional details on the activities of the compound. It includes Superkingdom, Kingdom, Phylum, Class, Order, Family, Genus, Species.
Taxonomies details: Taxonomies section shows the taxonomy tree of the compound. The depth of the tree can be controlled in the search panel, described above.
Patents: Similar to the Pubmed section, this section shows the patents associated with the compound. By clicking on the title of the patent, you will be redirected to the associated page. Again, the number on the right indicates the number of times that this compounds has been described in the patent. The three dots next to the number allow the user to see the abstract of the patent.
Obviously, all sections can be expanded and collapsed by clicking on the arrow on the right.
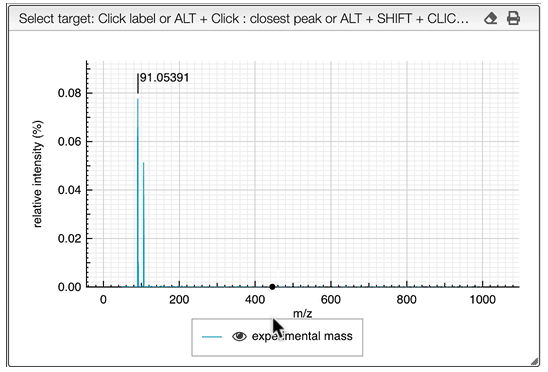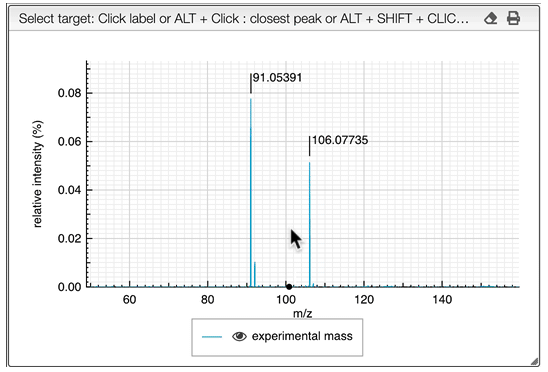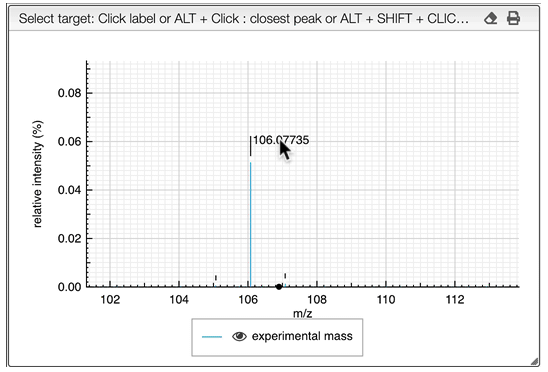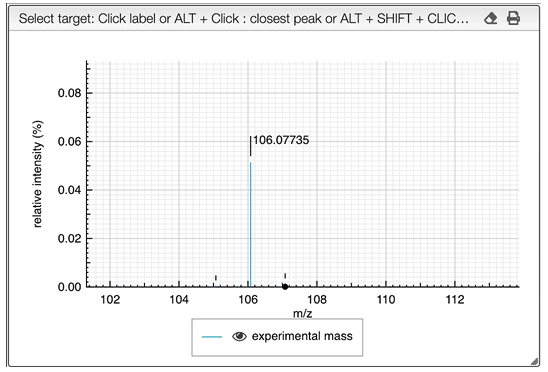
The other icon  appears when there are experimental spectra from the GNPS database. If you click on this icon, you will be able to compare your spectrum with spectra of molecules that have the same molecular formula from real experiments. You can change experimental spectra using the arrows in you keyboard.
appears when there are experimental spectra from the GNPS database. If you click on this icon, you will be able to compare your spectrum with spectra of molecules that have the same molecular formula from real experiments. You can change experimental spectra using the arrows in you keyboard.
In the table on the top you can see the list of available spectra. For each spectrum, there are information about the ion source, the instrument used to make the measure, the precursor, the ionization, the mode and the export icon redirects you to the original web page where the spectrum comes from. You can also see the similarity between your spectrum and the ones from the GNPS database. Your spectrum as well as the selected experimental spectra are plotted in a head to tail plot.
In the left text entry boxes, you can select the number of peaks that you want to see as well as the accuracy that you allow when comparing with the experimental spectrum from the database. The mass power and intensity power are a way to make either the mass or the intensity more important when comparing with the experimental spectrum. The higher is a power, the more important becomes the information (intensity or mass). The similarity between two spectra is calculated based on the cosine similarity.
On the bottom left panel, you can select any experimental spectrum that was imported.